MCCE Data Analysis
MCCE data analysis tools
fitpka.py
Fit the titration curve of an ionizable residue.
Syntax:
fitpka.py [-h] RES [RES ...]
Fit a titration of charged residues
positional arguments:
RES Charged residue names to plot, as in sum_crg.out or pK.out
optional arguments:
-h, --help show this help message and exit
Required input file
- sum_crg.out
Example:
Find the residue IDs:
$ cat sum_crg.out
pH 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14
NTR+A0001_ 1.00 1.00 1.00 1.00 0.99 0.96 0.70 0.20 0.03 0.00 0.00 0.00 0.00 0.00 0.00
LYS+A0001_ 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 0.97 0.78 0.27 0.04 0.00 0.00 0.00
ARG+A0005_ 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 0.97 0.80 0.38 0.08
GLU-A0007_ -0.00 -0.01 -0.07 -0.38 -0.84 -0.98 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00
LYS+A0013_ 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 0.99 0.95 0.67 0.19 0.03 0.00
ARG+A0014_ 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 0.99 0.94 0.66 0.19
HIS+A0015_ 1.00 1.00 1.00 1.00 1.00 0.98 0.83 0.36 0.06 0.01 0.00 0.00 0.00 0.00 0.00
ASP-A0018_ -0.01 -0.10 -0.49 -0.89 -0.99 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00
TYR-A0020_ -0.00 -0.00 -0.00 -0.00 -0.00 -0.00 -0.00 -0.00 -0.00 -0.00 -0.01 -0.03 -0.09 -0.32 -0.73
ARG+A0021_ 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 0.99 0.91 0.59 0.22
TYR-A0023_ -0.00 -0.00 -0.00 -0.00 -0.00 -0.00 -0.00 -0.00 -0.01 -0.06 -0.32 -0.76 -0.96 -0.99 -1.00
LYS+A0033_ 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 0.97 0.78 0.27 0.05 0.01 0.00
GLU-A0035_ -0.00 -0.00 -0.01 -0.03 -0.12 -0.46 -0.88 -0.98 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00
ARG+A0045_ 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 0.99 0.91 0.56 0.21
ASP-A0048_ -0.03 -0.21 -0.69 -0.95 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00
ASP-A0052_ -0.00 -0.01 -0.07 -0.44 -0.84 -0.95 -0.99 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00
TYR-A0053_ -0.00 -0.00 -0.00 -0.00 -0.00 -0.00 -0.00 -0.00 -0.00 -0.00 -0.00 -0.00 -0.00 -0.00 -0.00
ARG+A0061_ 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 0.98 0.90 0.57
ASP-A0066_ -0.03 -0.20 -0.68 -0.93 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00
ARG+A0068_ 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 0.99 0.95 0.76 0.34
ARG+A0073_ 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 0.98 0.85 0.39 0.09
ASP-A0087_ -0.01 -0.08 -0.44 -0.88 -0.98 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00
LYS+A0096_ 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 0.96 0.76 0.33 0.08 0.02 0.01
LYS+A0097_ 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 0.98 0.85 0.40 0.08 0.01 0.00
ASP-A0101_ -0.00 -0.00 -0.01 -0.09 -0.51 -0.91 -0.99 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00
ARG+A0112_ 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 0.98 0.87 0.43 0.09
ARG+A0114_ 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 0.99 0.94 0.63 0.17
LYS+A0116_ 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 0.97 0.75 0.27 0.05 0.01 0.00 0.00
ASP-A0119_ -0.00 -0.00 -0.03 -0.19 -0.71 -0.96 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00
ARG+A0125_ 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 0.99 0.90 0.55 0.13
ARG+A0128_ 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 0.99 0.97 0.79 0.31
CTR-A0129_ -0.00 -0.04 -0.29 -0.75 -0.96 -0.99 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00 -1.00
----------
Net_Charge 18.93 18.34 16.22 13.47 11.05 9.70 8.68 7.57 7.01 6.38 4.53 1.82 -0.64 -4.61 -9.32
Protons 18.93 18.34 16.22 13.47 11.05 9.70 8.68 7.57 7.01 6.38 4.53 1.82 -0.64 -4.61 -9.32
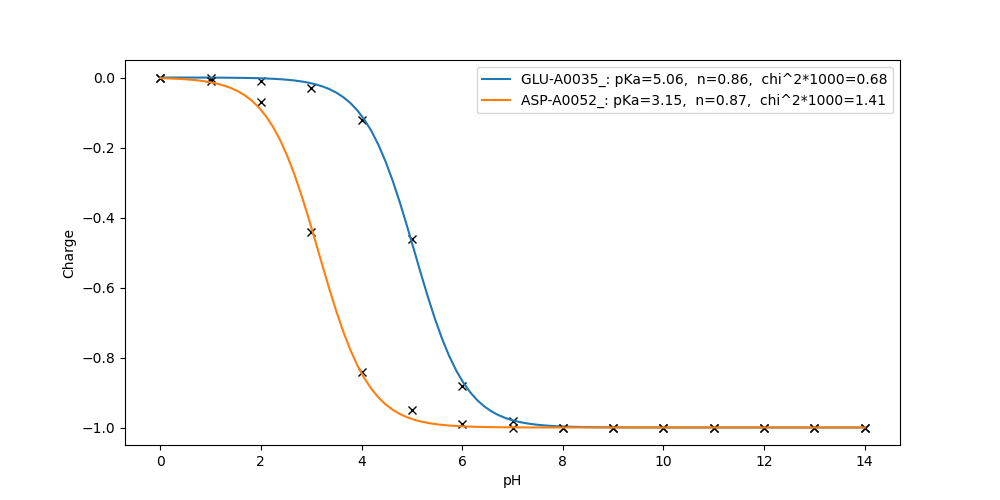
Electrons 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00Fit the titration curves of residue GLU-A0035_ and ASP-A0052_:
fitpka.py GLU-A0035_ ASP-A0052_
mfe.py
Analyze ionization free energy of a residue. It tells you why a residue has that pKa and what factors played a role.
Syntax
mfe.py [-h] [-p pH/Eh] [-x TS_correction] [-c cutoff] residue- residue: residue ID as in pK.out
- pH/Eh: pH value at which ionization energy is calculated. Default is the pKa (midpoint) where dG = 0.
- cut_off: Report only pairwise interaction bigger than this value.
- TS_correction: "t" to include entropy in G, "f" not to include, "r" (default) will look for run.prm. If entropy correction was turned on in step 4, mfe should not include this term as entropy has been "removed".
Required files:
- run.prm
- head3.lst
- extra.tpl for scaling factors that are not equal to 1.
- energies/*.opp for pairwise interactions
Example:
Check titration result in pKa.out:
cat pK.out
...
ASP-A0052_ 3.152
...
ASP-A0052_ is the residue_ID. Its calculated pKa is 3.152. At this point, the free energy of reaction from ASP neutral to ASP ionized should be close to 0.
Run
$ mfe.py ASP-A0052_ -c 0.1
Residue ASP-A0052_ pKa/Em=3.152
=================================
Terms pH meV Kcal
---------------------------------
vdw0 -0.01 -0.85 -0.02
vdw1 0.00 0.23 0.01
tors -0.10 -5.86 -0.14
ebkb -1.27 -73.44 -1.73
dsol 1.99 115.38 2.71
offset -0.62 -36.17 -0.85
pH&pK0 1.60 92.75 2.18
Eh&Em0 0.00 0.00 0.00
-TS 0.00 0.00 0.00
residues -1.36 -79.01 -1.86
*********************************
TOTAL 0.22 13.04 0.31 sum_crg
*********************************
ASNA0044_ -0.46 -26.92 -0.63 0.00
ARGA0045_ -0.11 -6.36 -0.15 1.00
ASNA0046_ -0.19 -11.09 -0.26 0.00
ASPA0048_ 0.50 28.96 0.68 -0.96
SERA0050_ 0.13 7.28 0.17 0.00
GLNA0057_ 0.22 12.78 0.30 0.00
ASNA0059_ -0.99 -57.18 -1.34 0.00
ARGA0061_ -0.26 -15.37 -0.36 1.00
ASPA0066_ 0.27 15.72 0.37 -0.94
ARGA0112_ -0.19 -11.11 -0.26 1.00
ARGA0114_ -0.10 -5.86 -0.14 1.00
=================================
You can do mfe calculation at pH other than mid-point.